MHCII-NP - Tutorial
How to obtain predictions
In order to do predictions, specify the sequences you want to scan for naturally processed MHC II ligands. The sequences should either be entered directly into the text area field or uploaded as a plain text file using the "Browse" button. Please enter no more than 100 FASTA sequences or upload file size more than 5MB per query. The sequences can be supplied as FASTA or plain format. The input will be interpreted as FASTA if an opening ">" character is found, or as a continuous sequence otherwise. All sequences have to be amino acids specified in single letter code (ACDEFGHIKLMNPQRSTVWY). Once sequences are uploaded submit them by clicking the "Submit" button.
Interpreting prediction output
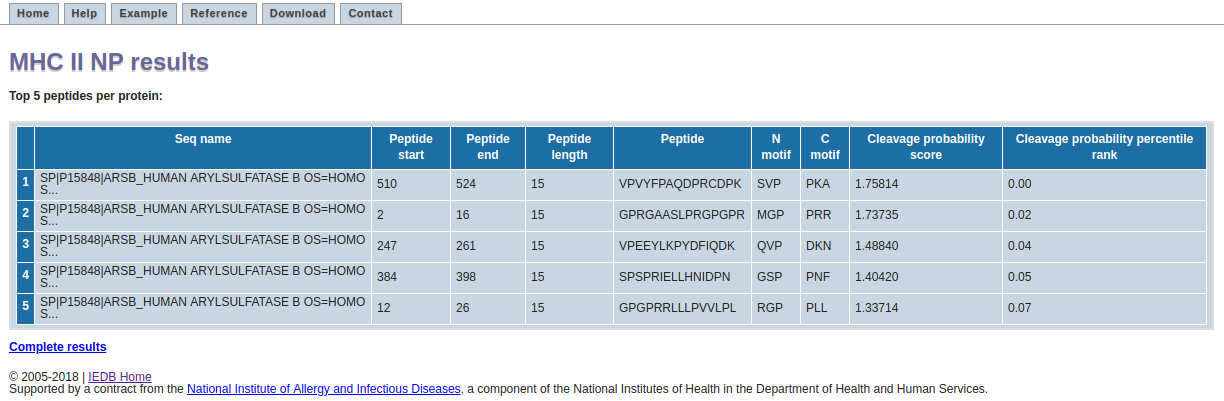
The result page displays top 5 peptides per each protein. Each row in the table corresponds to one prediction. The columns contain the sequence name, start and end positions of the peptide, its length, the peptide, the N- and C-terminuses considered for calculating the cleavage probability score, the predicted cleavage probability score and the predicted cleavage probability score in terms of percentile ranks per the corresponding source protein.
The higher the numerical value of cleavage probability score and the lower the numerical value of hte percentile score, the better is the chance of the peptide being a ligand.
The cleavage probability score indicates the probability of the peptide being an MHC II ligand based on three components: (1) the peptide length (2) the N-terminus cleavage motif and (3) the C-terminus cleavage motif. More details on the method can be obtained from the reference.
